Jun-Yong Choe
Associate Professor

- Email:
- jchoe@unm.edu
- Phone:
- 505 277-6655
- Office:
- Clark Hall 232
Research Interests:
- Chemical Biology
- Structural Biology
- X-ray crystallography
- Drug discovery
- Protein engineering
Education
- B.S. Biophysics, 1997, Iowa State University of Science and Technology
- M.S. Biochemistry, 1998, Iowa State University of Science and Technology
- Ph.D. Biophysics, 2001, Iowa State University of Science and Technology
- Post-doctoral Researcher, Chemistry, 2004, California Institute of Technology
Experience
- Senior Crystallographer, Structural Genomics Consortium, University of Toronto, (2004-2005)
- Assistant Research Physiologist, University of California Los Angeles, (2005-2007)
- Associate Professor / Assistant Professor, The Chicago Medical School, Rosalind Franklin University of Medicine and Science (2008-2018)
- Associate Professor, East Carolina Diabetes and Obesity Institute, East Carolina University (2018-2022)
Research
My main research interest is the structure-function relationship in carbohydrate-related and other proteins and its application to drug discovery, protein engineering and biotechnology. In our laboratory, we apply high-throughput methodologies for protein expression, functional analysis and crystallization. Besides x-ray protein crystallography, we routinely use membrane protein biochemistry, molecular biology and structural modeling.
Glucose transporters:
Carbohydrates serve as basic fuel molecules for most cells. These molecules are unable to freely diffuse across cellular membranes but instead require dedicated transporter proteins to either enter or exit cells. In human cells, the transport of glucose and related sugars is facilitated by a family of MFS transporters (SLC2 family) called glucose transporters (in short, GLUTs). As key players in the availability of cellular energy source, GLUTs have been involved in various diseases including diabetes, cancer and the metabolic syndrome. For example, cancer cells due to their higher energy demands compared to healthy cells often express higher levels of GLUT proteins: GLUT1 (a transporter of glucose) is overexpressed in most cancers, GLUT5 (a transporter of fructose) is increased in breast cancer though normal breast cells lack GLUT5, other GLUTs are specifically increased in various cancers. Inhibitors of GLUT proteins can deprive cancer cells of energy and thus become a powerful tool in curbing cancer, especially when combined with a way to target tumor cells. My main research focus is to determine the molecular basis of differences in the function of GLUT members and to uncover new GLUT ligands that are specific for a certain GLUT member in order to facilitate drug discovery in GLUT-related diseases.
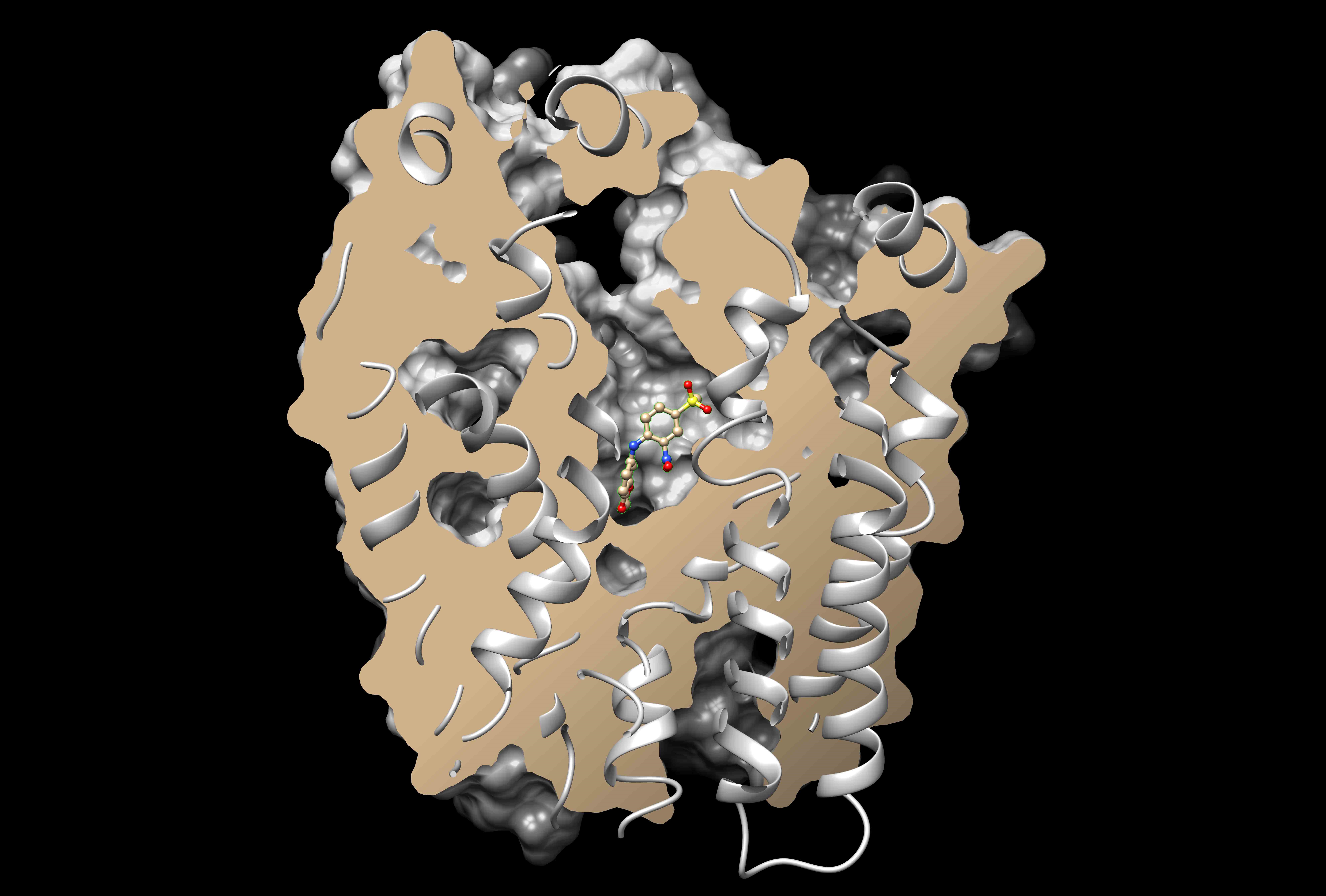
GLUT5-specific inhibitor, MSNBA, in the substrate binding cavity of GLUT5.
Salicylic acid storage in plants:
Salicylic acid (SA) is a plant hormone involved in regulating plant stress responses including local and systemic pathogen responses. SA is stored as a glucose conjugate in the form of either an SA glucoside (SAG) or an SA glucose ester (SGE), and these conjugation reactions are catalyzed by glucosyltransferases. In the model organism Arabidopsis thaliana the enzyme UGT74F1 forms SAG while UGT74F2 primarily forms SGE, however, the two enzymes share 90% similarity at the amino acid level. In collaboration with Dr. John Dean’s group from DePaul University, Chicago, through structural biology, biochemistry and molecular biology, we aim to understand how SA is processed and stored in the plant cell.
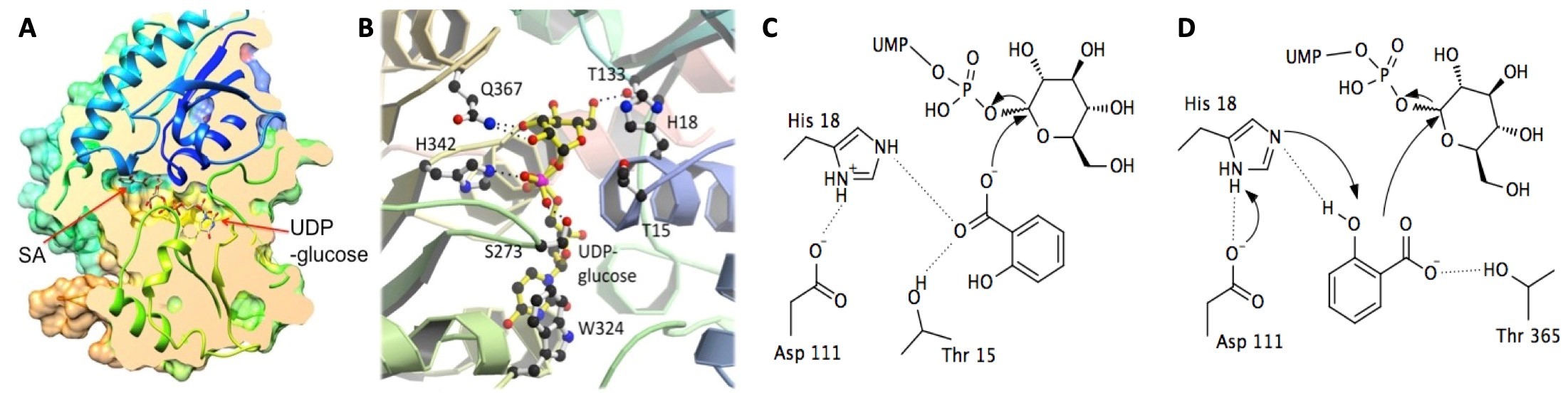
Crystal structures of UGT74F2. Overall structure with longitudinal section through the active site of UGT74F2 (A). Close-up of the active site showing binding site for UDP-glucose in UGT74F2 (B). The proposed catalytic mechanism for UGT74F1 (C) and UGT74F2 (D).
Co-factor independent decarboxylases:
Gallic acid decarboxylase catalyzes the formation of pyrogallol through the decarboxylation of gallic acid. Gallic acid, in turn, is a degradation product of tannins, polyphenolic biomolecules present in almost all types of plant tissues. Pyrogallol is very sensitive to oxygen and has many industrial applications. Our collaborator, Dr. Mislav Oreb (Johann Wolfgang Goethe-Universität Frankfurt am Main, Germany), is engineering yeast strains for biotechnological applications. We are working together on the structure and function of cofactor-free decarboxylases, which are easier to integrate into engineered pathways. We have solved the crystal structures of two fungal cofactor-free decarboxylases (AGDC1 from Arxula adenivorans, and PPP2 from Madurella mycetomatis), identified the active site using co-crystallization with an inhibitor, confirmed it by site-directed mutagenesis and mutant enzyme assays, and proposed the catalytic mechanism.
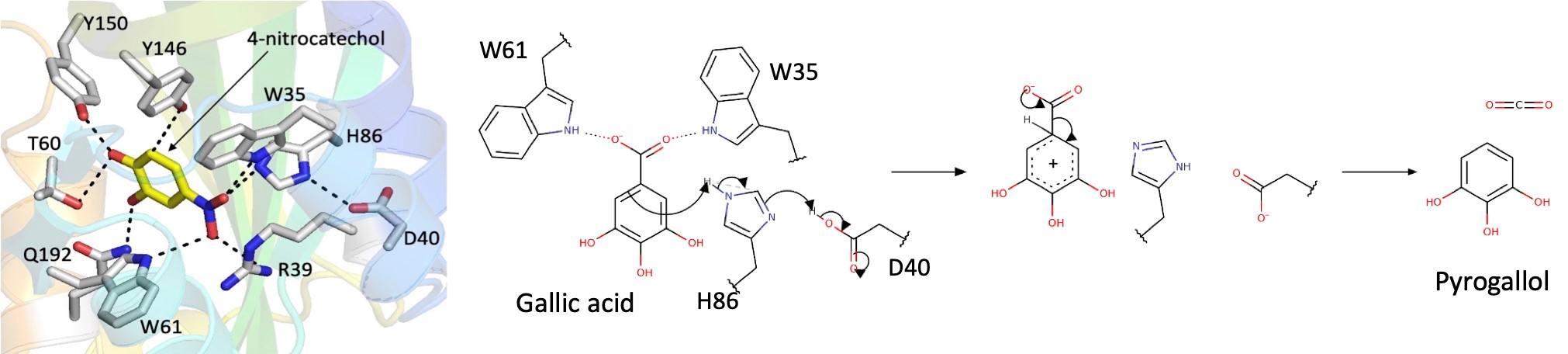
Cofactor-free proposed catalytic mechanism for gallic acid decarboxylase. A tryptophan clamp (W35 & W61) twists the carboxyl group increasing the energy of the substrate. H86 activated by D40 protonates gallate to form a Wheland intermediate. The reaction proceeds via an electrophilic aromatic substitution.
Recent Publications
Tyson, K., Tangtartharakul, C.B., Zeug, M., Findling, N., Haddy, A., Hvastkovs, E., Choe, J.Y., Kim, J.E., Offenbacher, A.R. (2023) “Electrochemical and Structural Study of the Buried Tryptophan in Azurin: Effects of Hydration and Polarity on the Redox Potential of W48.” Journal of Physical Chemistry B. 127(1):133-143.
Iancu, C.V., Bocci, G., Ishtikhar, M., Khamrai, M., Oreb, M., Oprea, I.T., Choe, J. (2022) “GLUT3 inhibitor discovery through in silico ligand screening and in vivo validation in eukaryotic expression systems.” Scientific Reports, 12, 1429.
Schmidl, S., Ursu, O., Iancu, C., Oreb, M, Oprea, T., Choe, J., (2021) “Identification of new GLUT2-selective inhibitors through in silico ligand screening and validation in eukaryotic expression systems.” Scientific Reports, 11, 13751.
Zeug, M., Markovic, N., Iancu, C.V., Tripp, J., Oreb, M, Choe, J. (2021) “Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.” Scientific Reports, 11, 3056.
Schmidl, S., Tamayo Rojas, S.A., Iancu, C.V. Choe, J., Oreb, M. (2021) “Functional expression of the human glucose transporters GLUT2 and GLUT3 in yeast offers novel screening systems for GLUT-targeting drugs.” Frontiers in Molecular Biosciences, 7, 598419.
Schmidl, S., Iancu, C., Reifenrath, M., Choe, J., Oreb, M. (2021) “A label-free real-time method for measuring glucose uptake kinetics.” FEMS Yeast Research, 21. foaa069.
Seiça, A.F.S., Iqbal, M.H., Carvalho, A., Choe, J., Hellwig, P. (2021) “Study of membrane proteins monolayers: modification of the surface properties during immobilization process using Surface-enhanced infrared absorption spectroscopy (SEIRAS)” ACS Sensors, 27, 2875.
Upadhyay, N., Tilekar, K., Loiodice, F., Yu, A.N., Tatiana, S., Darina., S., Galina, S., Choe, J., Meyer-Almes, J. Vadim S. P. Lavecchia, A., Ramaa C.S. (2021) “Pharmacophore Hybridization Approach to Discover Novel Pyrazoline-Based Hydantoin Analogs With Anti-tumor Efficacy.” Bioorganic Chemistry. 107, 104527.
Cheung, T., Choe, J., Richmond, J.E., Kim, H. (2020) “BK channel density is regulated by endoplasmic reticulum associated degradation and influenced by the SKN-1A/NRF1 transcription factor.” Plos Genetics 16, e1008829
Upadhyay, N., Tilekar, K., Jänsch, N., Schweipert, M., Jessica Hess, J., Henze, L., Mrowka, P., Aguilera, R., Choe, J., Meyer-Almes, J. (2020) “Discovery of novel N-substituted thiazolidinediones (TZDs) as HDAC8 inhibitors: in-silico studies, synthesis, and biological evaluation.” Bioorganic Chemistry 100, 103934.
Tilekar, K., Upadhyay, N., Hess, J.D., Macias, L.H., Mrowka, P., Renato, A., Meyer-Almes, F., Iancu, C.V., Choe, J., Ramaa, C.S, (2020) “Design and synthesis of furyl thiazolidinedione derivatives as inhibitors of GLUT1 and GLUT4 and evaluation of their in-vitro and in-vivo anti-leukemic potential.” European Journal of Medicinal Chemistry 202, 112603.
Tilekar, K., Upadhyay, N., Schweipert M, Hess J.D, Macias L.H, Mrowka P, Meyer-Almes F.J, Aguilera R.J, Iancu C.V, Choe J, Ramaa C.S (2020) “Permuted 2,4-thiazolidinedione (TZD) analogs as GLUT inhibitors and their in-vitro evaluation in leukemic cells” European Journal of Pharmaceutical Science 154, 105512.
Santos Seica, A.F., Iancu, C.V., Pfeilschifter, B., Madej, M.G., Choe, J., Hellwig, P. (2020) “Asp22 drives the protonation state of the Staphylococcus epidermidis glucose/H + symporter” J. Biol Chem. 295, 15253-61.
Tilekar, K., Upadhyay, N., Iancu C.V, Pokrovsky, V., Choe J, Ramaa C.S (2020) “Power of two: combination of therapeutic approaches involving glucose transporter (GLUT) inhibitors to combat cancer” Biochimica et Biophysica Acta (BBA) – Reviews on Cancer, 1874, 188457.
Harth, S., Wagner, J., Sens, T., Choe, J., Benz, J.P., Weuster-Botz, D., Oreb, M. (2020) “Engineering cofactor supply and NADH-dependent D-galacturonic acid reductases for redox-balanced production of L-galactonate in Saccharomyces cerevisiae” Scientific Reports, 10, 19021.
Behrens C.E, Smith K.E, Iancu C.V, Choe J, Dean J.V. (2019) “Transport of Anthocyanins and other Flavonoids by the Arabidopsis ATP-Binding Cassette Transporter AtABCC2.” Scientific Reports 9, 437.
Ferraris, R.P., Choe, J., and Patel, C.R. (2018) “Intestinal Absorption of Fructose.” Annual Review of Nutrition. 38, 41-67.
Culbertson, A.T., Ehrlich, J.J., Choe, J., Honzatko, R.B., and Zabotina, O.A. (2018) “Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.” Proc. Natl. Acad. Sci. U. S. A. 115, 6064-6069.
Schmidl, S., Iancu, C.V., Choe, J., and Oreb, M. (2018) “Ligand Screening Systems for Human Glucose Transporters as Tools in Drug Discovery” Frontiers in Chemistry. 6, 183.
George Thompson, A.M., Iancu, C.V., Neet, K.E., Dean, J.V., and Choe, J. (2017) “Differences in substrate binding mode and catalytic mechanism lead to distinct salicylic acid glucose conjugates by UGT74F1 and UGT74F2 from Arabidopsis thaliana.” Scientific Reports 7, 46629.
Thomik, T., Wittig, I., Choe, J., Boles, E., and Oreb, M. (2017) “An artificial transport metabolon facilitates improved substrate utilization in yeast.” Nature Chemical Biology. 13, 1158-1163.
Vaca, E., Behrens, C., Theccanat, T., Choe, J., and Dean, J.V. (2017) “Mechanistic differences in the uptake of salicylic acid glucose conjugates by vacuolar membrane-enriched vesicles isolated from Arabidopsis thaliana.” Plant Cell Physiol. 161, 322-338.
Tripp, J., Essl, C., Iancu, C., Boles, E. Choe, J., and Oreb, M. (July 2017) “Establishing a yeast-based screening system for discovery of human GLUT5 inhibitors and activators.” Scientific Reports 7, 6197
George Thompson, A.M., Ursu, O., Iancu, C.V., Babkin, P, Oprea, T.I., and Choe, J. (2016) “Discovery of a specific inhibitor of human GLUT5 by virtual screening and in vitro transport evaluation.” Scientific Reports 6, 24240.
Jordan, P., Choe, J., Boles, E., and Oreb, M. (2016) “Hxt13, Hxt15, Hxt16 and Hxt17 of Saccharomyces cerevisiae represent a novel type of polyol transporters.” Scientific Reports 6, 23502.
Milton, M.E., Choe, J., Honzatko, R.B., Nelson, S.W. (2016) “Crystal structure of the apicoplast DNA polymerase from Plasmodium falciparum: the first look at an “atypical” A-family DNA polymerases.” Journal of Molecular Biology 428, 3920-3934.
Mandal, T., Shin, S., Aluvila, S., Chen, H.C., Grieve, C., Choe, J., Cheng, E. H., Hustedt, E. J., and Oh, K. J. (2016) “Assembly of Bak homodimers into higher order homooligomers in the mitochondrial apoptotic pore.” Scientific Reports 6, 30763.
Milton, M.E., Choe, J., Honzatko, R.B., Nelson, S.W. (2015) “Crystallization and preliminary X-ray analysis of the Plasmodium falciparum apicoplast DNA polymerase.” Acta Crystallogr. F Struct. Biol. Commun. 71, 333-337.
Babkin, P, George Thompson, A.M., Iancu, C.V., Walters, D.E., and Choe, J. (2015) “Antipsychotics inhibit glucose transport: determination of olanzapine binding site in Staphylococcus epidermidis glucose/H+ symporter.” FEBS OpenBio 5, 335–340.
George Thompson, A.M., Iancu, C.V., Nguyen, T.T.H. Kim, D., and Choe, J. (2015) “Inhibition of human GLUT1 and GLUT5 by plant carbohydrate products; insights into transport specificity.” Scientific Reports 5, 12804.
Choe, J. (2015) “Crystallography as Art: Revealing the Inner Workings of a Glucose Transporter.” Acad. Medicine 90, 1631.
Aluvila, S., Mandal, T., Hustedt, E., Fajer, P., Choe, J., Oh, K.J. (2014) “Organization of the Mitochondrial Apoptotic BAK Pore: Oligomerization of the BAK Homodimers.” J. Biol. Chem. 289, 2537-2551.
Gao, Y., Iancu, C.V., Mukind, S., Choe, J., and Honzatko, R.B. (2013) “Mechanism of Displacement of a Catalytically Essential Loop from the Active Site of Mammalian Fructose-1,6-bisphosphatase.” Biochemistry 52, 5206-5216.
Iancu, C.V., Zamoon, J., Woo, S.B., Aleshin, A., and Choe, J. (2013) “Crystal structure of a glucose/H+ symporter and its mechanism of action.” Proc Natl Acad Sci U S A 110, 17862–17867.
